Bioinformatics group
at Warsaw University of Life Sciences
Proteins, the basic building blocks and machines that make all living cells function, are fascinating molecules. Many of them have been meticulously studied for decades, but others are completely uncharacterised.
If you don’t know what your favourite protein is doing, we may be able to find the answer, or, at least, suggest the kind of experiments that may help find the answer.
If you ever wondered whether your favourite protein family/superfamily is fully charted or if it may have distant cousins hitherto unidentified, we may be able to find out.
We use classical sequence and structure bioinformatics approaches, data mining, and anything that works or may work to cast light on possible functions of the zillions of uncharacterised proteins.
We also map high-throughput biological data, primarily proteomics, on biological molecular relationships and clinical parameters to gain insight into disease mechanisms.

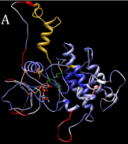
Structure and function predictions for uncharacterised proteins potentially important in biology and in disease
![]()
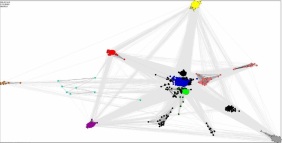
Novel gene/protein families with unexpected evolutionary histories
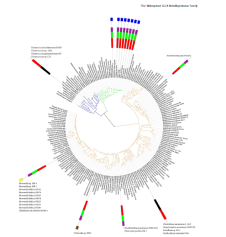
Systems biology analysis of high throughput biological data
Current projects
- Search for novel protein kinase-like effectors in bacterial pathogens and throughout the tree of life; structure and function predictions
- In silico search for, and characterisation of, novel enzyme families; focus on kinases, ADP-ribosyltransferases, acetyltransferases, proteases, oxidoreductases
- Development of bioinformatics tools for functional analyses of genomic neighborhoods and for gene co-occurrence analysis
- Systems biology analyses of biomedical proteomics data

You must be logged in to post a comment.